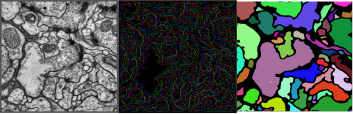
Neuron segmentation in EM images using local constraint based integer linear programming
Methods for analyzing neuroanatomy using electron microscopy images
Since the advent of electron microscopy (EM) in the early 1950s, it has been a widely used tool to answer many questions in neuroscience due to its ability to acquire images at very high resolutions that enabled scientists to see neuronal ultra-structures as small as synapses and vesicles filled with neurotransmitters.
Different imaging modalities of EM such as serial section Scanning Electron Microscopy (ssSEM), Focused Ion Beam Scanning Electron Microscopy (FIBSEM) are used to not only to study general structural properties of neural systems, but also to detect changes in such structures due to specific stimuli (e.g. tutoring of juvenile song birds). Most of these studies require EM images to be annotated with the relevant labels of neuronal structures (neuron sections, synapses, mitochondria, myelin sheaths, vesicles). This project focuses on developing image processing methods for automatic annotation of such structures in EM images and using both automatic and manual annotation schemes to study neuroanatomy. We then use these methods to investigate neuroanatomical changes caused by experimental stimuli.