Spiking autoencoder for muscle synergies extraction from EMG data- Semester/ Master thesis project
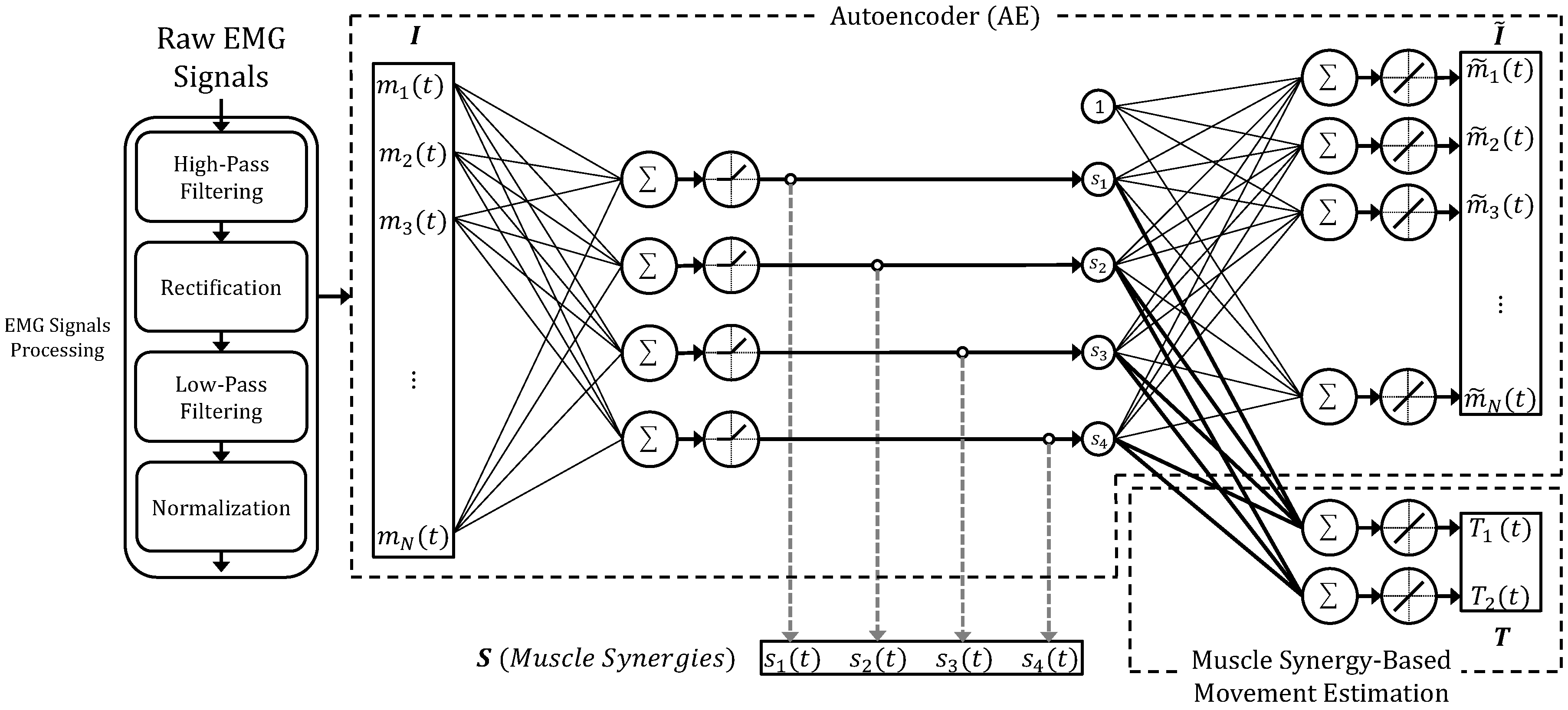
Figure 1 Example of Autoencoder-based neural model, proposed in [4]
The Institute of Neuroinformatics (INI) is a joint institute of the University of Zurich and the Swiss Federal Institute of Zurich (ETH Zurich). INI carries out experimental, theoretical and applied research with the aim of discovering key principles by which the brain is built and works and using this knowledge in practical applications where possible. The Neuromorphic Cognitive Systems (NCS) group at INI has been developing neuromorphic processing platforms that enable event-driven computing using algorithms and primitives inspired by biology.
An autoencoder is a particular type of neural network that is trained to replicate its input. For example, given an image of a handwritten digit, an autoencoder first encodes the image into a lower dimensional latent representation, then decodes the latent representation back to an image. An autoencoder learns to compress the data while minimizing the reconstruction error. As such they can be deployed to learn the representation of any signal, including bio-signals. In particular, in EMG processing, autoencoders are used to map EMG signals into kinematics [1], for joint compression and classification [2] or for extracting information in the latent space [3,4].
In this project the candidate will explore a spiking implementation of an autoencoder to extract motor intent (muscle synergies). Creating a spiking neural network (SNN) which is able to process data and keep memory of past inputs is an open challenge in the field. The candidate will have the opportunity to learn about SNNs, learning rules, and interfacing networks with real world signals. The project includes a set of intermediate milestones such as, understanding network topology, training the SNN and validating the network. If time allows (probably for master thesis) the candidate could interface the network created with a robotic hand.
Reference:
[1] Vujaklija, I., Shalchyan, V., Kamavuako, E. N., Jiang, N., Marateb, H. R., & Farina, D. (2018). Online mapping of EMG signals into kinematics by autoencoding. Journal of neuroengineering and rehabilitation, 15(1), 1-9.
[2] Said, A. B., Mohamed, A., Elfouly, T., Harras, K., & Wang, Z. J. (2017, March). Multimodal deep learning approach for joint EEG-EMG data compression and classification. In 2017 IEEE Wireless Communications and Networking Conference (WCNC) (pp. 1-6). IEEE. Chicago
[3] Zia ur Rehman, M., Waris, A., Gilani, S. O., Jochumsen, M., Niazi, I. K., Jamil, M., ... & Kamavuako, E. N. (2018). Multiday EMG-based classification of hand motions with deep learning techniques. Sensors, 18(8), 2497.
[4] Buongiorno, D., Cascarano, G. D., Camardella, C., De Feudis, I., Frisoli, A., & Bevilacqua, V. (2020). Task-Oriented Muscle Synergy Extraction Using An Autoencoder-Based Neural Model. Information, 11(4), 219. Chicago
Requirements
Programming skills in Python,
Interest in neuronal systems (biological or artificial).
Start of project: Immediately
Length of project: Master Thesis/Semester project
Contact
Elisa Donati: elisa (at) ini.uzh.ch
Matteo Cartiglia: camatteo (at) ini.uzh.ch