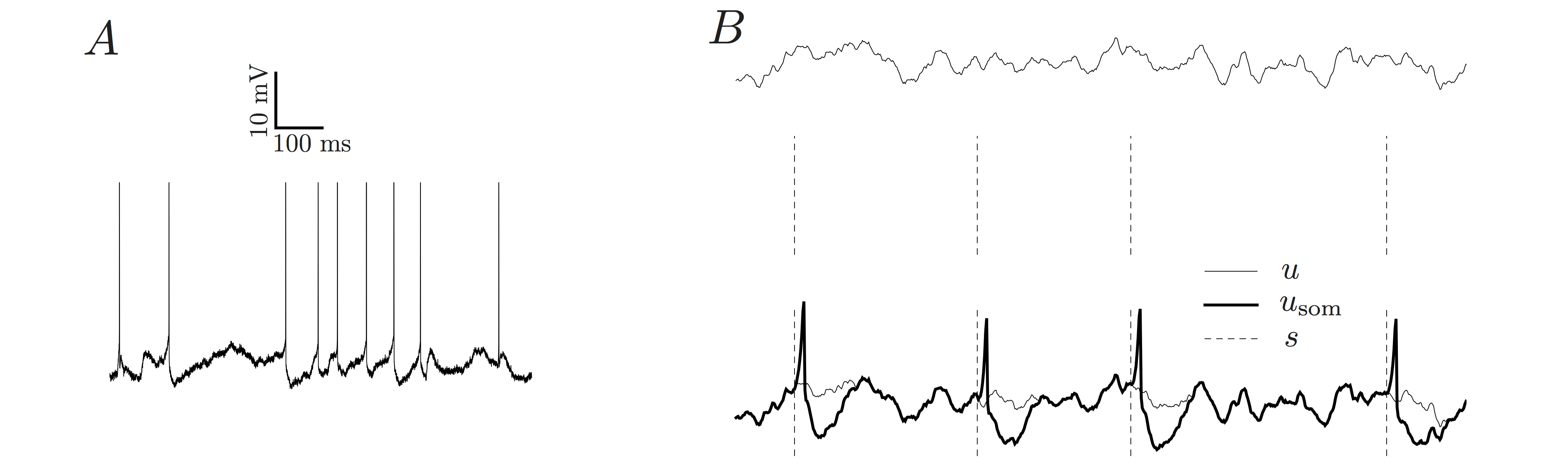
(A) A sample in vivo membrane potential trace from an intracellular recording of a neuron in HVC of a Zebra Finch. (B) The generative AGAPE model can generate a trace of subthreshold membrane potential u (top trace). Based on this potential, a spike train s is generated (middle, dashed vertical lines). Finally, a stereotypic spike-related kernel is convolved with the spike train, giving rise to usom (bottom, thick line). This quantity is the synthetic analog of the recorded, preprocessed in vivo membrane potential.
Statistical model for intracellular in vivo recordings
Single neuron models have a long tradition in computational neuroscience. Detailed biophysical models such as the Hodgkin-Huxley model as well as simplified neuron models such as the class of integrate- and-fire models relate the input current to the membrane potential of the neuron. Those types of model have been extensively fitted to in vitro data where the input current is controlled. Those models are however of little use when it comes to characterize intracellular in vivo recordings since the input to the neuron is not known. Here we propose a novel single neuron model that characterizes the statistical properties of in vivo recordings. More specifically, we propose a stochastic process where the subthreshold membrane potential follows a Gaussian process and the spike emission intensity depends nonlinearly on the membrane potential as well as the previous spiking history. We first show that the model has a rich dynamical repertoire since it can capture arbitrary subthreshold autocovariance functions, firing- rate adaptations as well as arbitrary shapes of the action potential. We then show that this model can be efficiently fitted to data without overfitting. We finally show that this model can be used to characterize and therefore precisely compare various intracellular in vivo recordings from different animals and recording conditions.